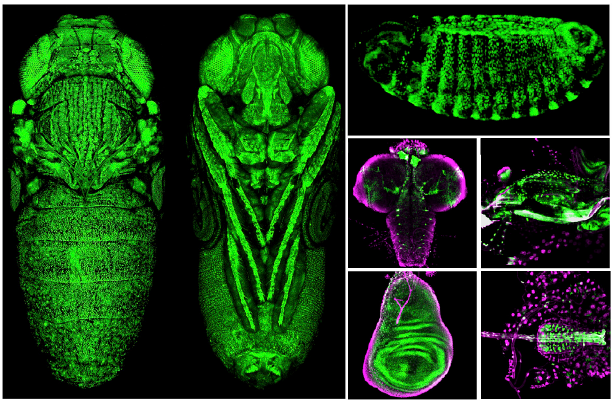
Architecture and Evolution of Regulatory DNA
Although a wealth of genomic information is deposited in databases, our understanding of the function and evolution of regulatory DNA remains superficial. We are only beginning to comprehend how regulatory information is distributed within non-coding DNA, how regulatory elements interact, how a single regulatory element functions in different organs, and how regulatory regions evolve across species. Our goal is to gain insights into the structure and evolution of the DNA that regulates gene activity. Currently, we focus on the study of pleiotropic enhancers—elements that encode regulatory information for generating more than one expression pattern. We aim to explore the extent of pleiotropy in regulatory regions and to uncover the molecular mechanisms that modulate the activity of pleiotropic enhancers in different developmental contexts.
We use the Drosophila gene shavenbaby (svb) to investigate mechanistic and evolutionary aspects of gene regulation. svb encodes a master transcription factor that governs the development of trichomes (nonsensory cuticular hairs) in both larvae and adults. Seven transcriptional enhancers distributed across the regulatory region of svb control its embryonic expression. Each embryonic enhancer functions in a distinct group of cells, although there is some redundancy among the expression patterns driven by these enhancers. In the pupal epidermis, the regulatory architecture underlying svb expression appears to be even more complex than in the embryo. Regulatory DNA driving pupal epidermal expression spans large genomic regions that are inactive during embryogenesis. Expression patterns driven by DNA fragments within these regions are highly redundant in the pupal abdominal epidermis. We aim to understand the mechanisms underlying svb activation in the embryo and pupa, and why these mechanisms differ between developmental stages.
Li XC, Srinivasan V, Laiker I, Misunou N, Frankel N, Pallares LF, Crocker J. TF-High-Evolutionary: in vivo mutagenesis of gene regulatory networks for the study of the genetics and evolution of the Drosophila regulatory genome. Molecular Biology & Evolution (2024) 41 (8): msae167.
Sabarís G, Ortíz DM, Laiker I, Mayansky I, Naik S, Cavalli G, Stern DL, Preger-Ben Noon E, Frankel N. The Density of Regulatory Information Is a Major Determinant of Evolutionary Constraint on Noncoding DNA in Drosophila. Molecular Biology & Evolution (2024) 41(2): msae004.
Moreyra NN, Almeida FC, Allan C, Frankel N, Matzkin LM, Hasson E. Phylogenomics provides insights into the evolution of cactophily and host plant shifts in Drosophila. Molecular Phylogenetics & Evolution (2023)178:107653.
Preger-Ben Noon E, Frankel N. Can changes in 3D genome architecture create new regulatory landscapes that contribute to phenotypic evolution? Essays in Biochemistry (2022) 66(6):745-752.
Laiker I, Frankel N. Pleiotropic enhancers are ubiquitous regulatory elements in the human genome. Genome Biology and Evolution (2022) 14(6), evac071.
Kittelmann S, Preger-Ben Noon E, McGregor AP, Frankel N. A complex gene regulatory architecture underlies the development and evolution of cuticle morphology in Drosophila. Current Opinion in Genetics and Development (2021) 69:21-27.
Soverna AF, Rodriguez NC, Korgaonkar A, Hasson E, Stern DL, Frankel N. Cis-regulatory variation in the shavenbaby gene underlies intraspecific phenotypic variation, mirroring interspecific divergence in the same trait. Evolution (2021) 75:427-436.
Saraceno M, Gómez Lugo S, Ortiz N, Gómez BM, Sabio y Garcia CA, Frankel N, Graziano M.Unraveling the ecological processes modulating the population structure of Escherichia coli in a highly polluted urban stream network. Scientific Reports (2021) 11: 14679.
Sabarís G, Laiker I, Preger-Ben Noon E, Frankel N. Actors with Multiple Roles: Pleiotropic Enhancers and the Paradigm of Enhancer Modularity. Trends in Genetics (2019) 35:423‐433.
Kittelmann S, Buffry AD, Franke FA, Almudi I, Yoth M, Sabaris G, Couso JP, Nunes MDS, Frankel N, Gómez-Skarmeta JL, Pueyo-Marques J, Arif S, McGregor AP. Gene regulatory network architecture in different developmental contexts influences the genetic basis of morphological evolution. PLoS Genetics (2018) 14:e1007375.
Preger-Ben Noon E*, Sabarís G*, Ortiz DM, Sager J, Liebowitz A, Stern DL, Frankel N. Comprehensive Analysis of a cis-Regulatory Region Reveals Pleiotropy in Enhancer Function. Cell Reports (2018) 22:3021-3031.
Stern DL, Crocker J, Ding Y, Frankel N, Kappes G, Kim E, Kuzmickas R, Lemire A, Mast JD, Picard S. Genetic and Transgenic Reagents for Drosophila simulans, D. mauritiana, D. yakuba, D. santomea, and D. virilis. G3 (2017) 7:1339-1347.
Colines B, Rodríguez NC, Hasson ER, Carreira V, Frankel N. Parental age influences developmental stability of the progeny in Drosophila. Proceedings of the Royal Society B (2015) 282. pii: 20142437.
Preger-Ben Noon E, Frankel N. Evolving genital structures: a deep look at network co-option. Developmental Cell (2015). 34:485-6
Crocker J, Abe N, Rinaldi L, McGregor AP, Frankel N, Wang S, Alsawadi A, Valenti P, Plaza S, Payre F, Mann RS, Stern DL. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell (2015)160:191-203.
Stern DL, Frankel N. The structure and evolution of cis-regulatory regions: the shavenbaby story. Philosophical Transactions of the Royal Society B (2013) 368:20130028.
Domínguez PG, Frankel N, Mazuch J, Balbo I, Iusem N, Fernie AR, Carrari F. ASR1 mediates glucose-hormone crosstalk by affecting sugar trafficking in tobacco plants. Plant Physiology (2013) 161:1486-500.
Frankel N, Wang S, Stern DL. Conserved regulatory architecture underlies parallel genetic changes and convergent phenotypic evolution. Proceedings of the National Academy of Sciences USA (PNAS) (2012) 109(51):20975-9.
Frankel N. Multiple layers of complexity in cis-regulatory regions of developmental genes. Developmental Dynamics (2012) 241:1857-66.
Frankel N*, Erezyilmaz DF*, McGregor AP, Wang S, Payre F, Stern DL. Morphological evolution caused by many subtle-effect substitutions in a transcriptional enhancer. Nature (2011) 474:598–603.
Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature (2010) 466:490-493.
Group members

Lic. Ignacio Mayansky
PhD student

Lic. Ignacio Mayansky
PhD student

Lic. Ian Laiker
PhD student


Lic. Ailen Altamirano
PhD student

Lic. Ailen Altamirano
PhD student
- Phone:+549 (11) 2546-6589
- Email:info@example.com

Lic. Nicolas Portnoy
PhD student

Lic. Nicolas Portnoy
PhD student
- Phone:+549 (11) 2546-6589
- Email:info@example.com
Sofía Restuccia
Undergraduate
Sofía Restuccia
Undergraduate
- Phone:+549 (11) 2546-6589
- Email:info@example.com
Julián Rodriguez
Undergraduate
Julián Rodriguez
Undergraduate
- Phone:+549 (11) 2546-6589
- Email:info@example.com
Alumni
Dr. Gonzalo Sabarís
Dr. Daniela Ortiz
Dr. Martin Saraceno